INTRODUCTION
In this blog, we will explore the mushroom dataset and use a Naive Bayes classifier to predict the edibility of mushrooms.
Importing Necessary Packages:
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
Reading the data:
mush=pd.read_csv('mushrooms.csv')
Exploratory Data Analysis(EDA):
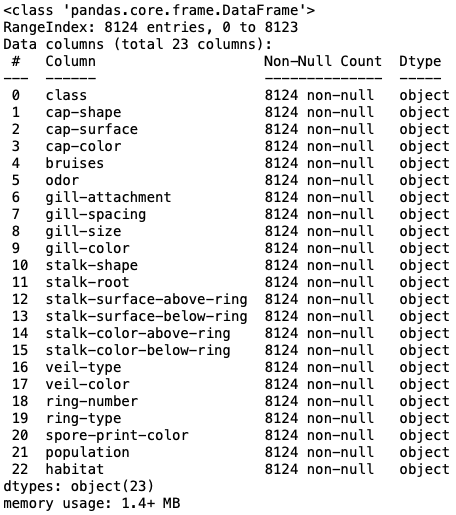
mush.info()
mush.columns

mush.isna().sum()
Statistical Inference:
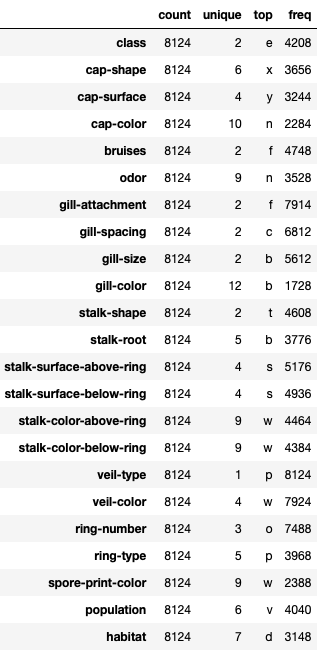
mush.describe().T
mush['class'].value_counts(normalize=True)
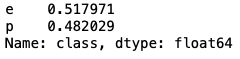
The mushroom dataset is fairly balanced in nature.
Here, we will build a Naive Bayes classifier to predict whether the mushroom is 'e' for edible or 'p' for poisonous.
Looking after the unique values in each columns of mushroom dataset:
for i in mush.columns:
print(i,mush[i].unique())

mush['stalk-root'].value_counts(normalize=True)
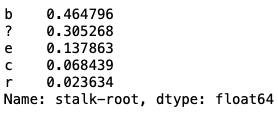
We can observe that the 'stalk-root' is a single column with '?' present in around 30% of its entries. Since 30% of the data represents a significant amount and cannot be removed from each column, the alternative is to remove the entire 'stalk-root' column.
Removing the stalk-root:
mush=mush.drop(columns=['stalk-root'])
mush.info()
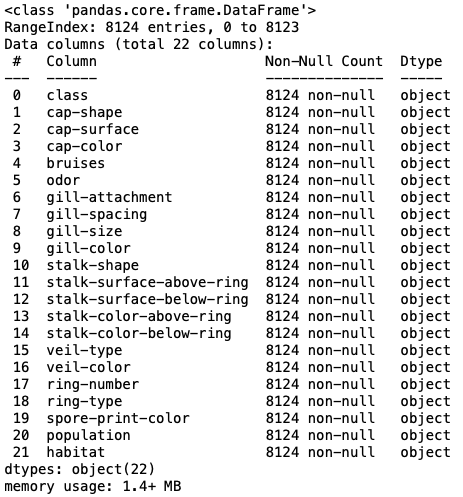 The column-stalk root is removed.
The column-stalk root is removed.
Data Preparation:
#Input Features:
X=mush.iloc[:,1:]
print(X)
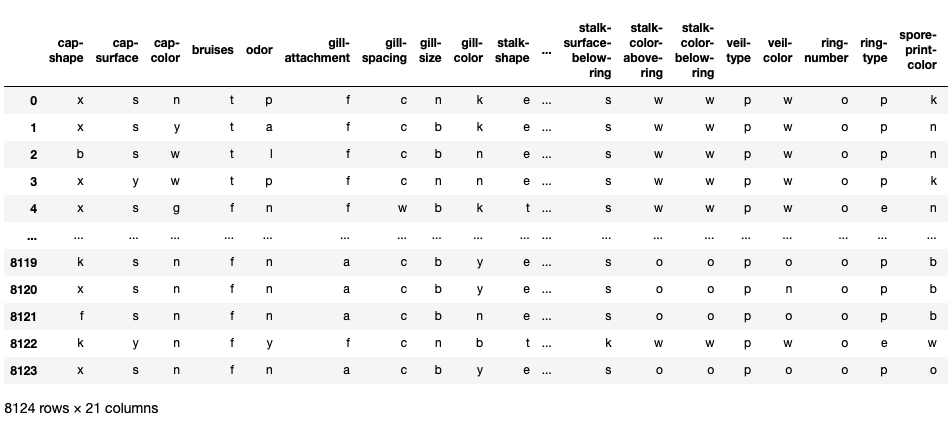
#Output:
Y=mush['class']
print(Y)

Since the inputs are categorical in nature, we need to convert them into numerical variables, as the Naive Bayes classifier deals with probability. We can achieve this conversion using 'get_dummies/Label Encoding.
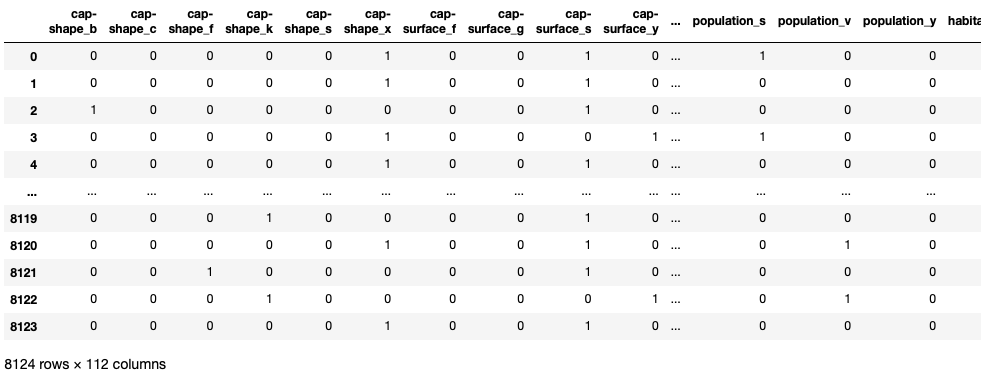
X=pd.get_dummies(X)
The 'get_dummies' function operates by taking a DataFrame, series, or list and transforming each distinct element into a column header, subsequently assigning a value of 1 if there's a match and 0 if there isn't.
print(X)
Splitting the data into training and testing:
from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test=train_test_split(X,Y,test_size=0.3,random_state=1)
print(x_train.shape)
print(x_test.shape)
print(y_train.shape)
print(y_test.shape)
(5686, 112)
(2438, 112)
(5686,)
(2438,)
Training the model:
from sklearn.naive_bayes import GaussianNB
NB_classifier=GaussianNB()
NB_classifier.fit(x_train,y_train)
Predicting the output:
y_pred=NB_classifier.predict(x_test)
Model Evaluation:
from sklearn.metrics import classification_report,accuracy_score,confusion_matrix
print(classification_report(y_test,y_pred))
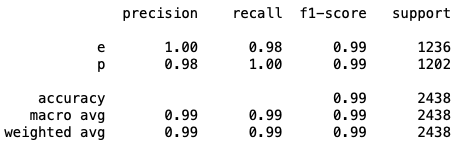
print(confusion_matrix(y_test,y_pred))
[[1216 20]
[ 2 1200]]
print(accuracy_score(y_test,y_pred))
0.9909762100082035
Conclusion:
This concludes our notebook. We used a Naive Bayes classifier from the scratch, trained it on our dataset, and observed an impressive accuracy of 99% during testing.
